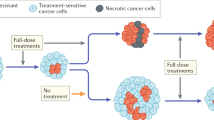
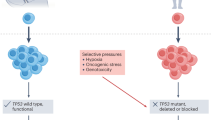
Abstract
Most targeted anticancer therapies fail due to drug resistance evolution. Here we show that tumor evolution can be reproducibly redirected to engineer therapeutic opportunity, regardless of the exact ensemble of pre-existing genetic heterogeneity. We develop a selection gene drive system that is stably introduced into cancer cells and is composed of two genes, or switches, that couple an inducible fitness advantage with a shared fitness cost. Using stochastic models of evolutionary dynamics, we identify the design criteria for selection gene drives. We then build prototypes that harness the selective pressure of multiple approved tyrosine kinase inhibitors and employ therapeutic mechanisms as diverse as prodrug catalysis and immune activity induction. We show that selection gene drives can eradicate diverse forms of genetic resistance in vitro. Finally, we demonstrate that model-informed switch engagement effectively targets pre-existing resistance in mouse models of solid tumors. These results establish selection gene drives as a powerful framework for evolution-guided anticancer therapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Sequencing data associated with this work are publicly available on the NIH NCBI SRA (BioProject PRJNA1081395)77. All other data are available in the main text, in Supplementary Information, or on GitHub in the associated figure directory at https://github.com/pritchardlabatpsu/SelectionGeneDrives74. Source data are provided with this paper.
Code availability
All code associated with this work is publicly available on GitHub (https://github.com/pritchardlabatpsu/SelectionGeneDrives)74.
References
Navin, N. E. The first five years of single-cell cancer genomics and beyond. Genome Res. 25, 1499–1507 (2015).
Schmitt, M. W. et al. Single-molecule sequencing reveals patterns of pre-existing drug resistance that suggest treatment strategies in Philadelphia-positive leukemias. Clin. Cancer Res. 24, 5321–5334 (2018).
Frankell, A. M. et al. The evolution of lung cancer and impact of subclonal selection in TRACERx. Nature 616, 525–533 (2023).
Martínez-Ruiz, C. et al. Genomic–transcriptomic evolution in lung cancer and metastasis. Nature 616, 543–552 (2023).
Song, P. et al. Limitations and opportunities of technologies for the analysis of cell-free DNA in cancer diagnostics. Nat. Biomed. Eng. 6, 232–245 (2022).
Short, N. J. et al. Ultra-accurate duplex sequencing for the assessment of pretreatment ABL1 kinase domain mutations in Ph+ ALL. Blood Cancer J. 10, 1–9 (2020).
Leighow, S. M. & Pritchard, J. R. The risks of perpetuating an evolutionary arms race in drug discovery. Evol. Med. Public Health 2019, 64–65 (2019).
Leonetti, A. et al. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer 121, 725–737 (2019).
Bozic, I. et al. Evolutionary dynamics of cancer in response to targeted combination therapy. eLife 2, e00747 (2013).
Goldie, J. H. & Coldman, A. J. The genetic origin of drug resistance in neoplasms: implications for systemic therapy. Cancer Res. 44, 3643–3653 (1984).
Frei, E. Curative cancer chemotherapy. Cancer Res. 45, 6523–6537 (1985).
Palmer, A. C., Chidley, C. & Sorger, P. K. A curative combination cancer therapy achieves high fractional cell killing through low cross-resistance and drug additivity. eLife 8, e50036 (2019).
Behan, F. M. et al. Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature 568, 511–516 (2019).
Meyers, R. M. et al. Computational correction of copy-number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017).
US Food and Drug Administration. FDA Approves Osimertinib with Chemotherapy for EGFR-Mutated Non-small Cell Lung Cancer (FDA, 2024).
Corrie, P. G. Cytotoxic chemotherapy: clinical aspects. Medicine 36, 24–28 (2008).
Wang, Y. et al. Clinical effectiveness and clinical toxicity associated with platinum-based doublets in the first-line setting for advanced non-squamous non-small cell lung cancer in Chinese patients: a retrospective cohort study. BMC Cancer 14, 940 (2014).
Noronha, V. et al. Gefitinib versus gefitinib plus pemetrexed and carboplatin chemotherapy in EGFR-mutated lung cancer. J. Clin. Oncol. 38, 124–136 (2020).
Planchard, D. et al. Osimertinib with or without chemotherapy in EGFR-mutated advanced NSCLC. N. Engl. J. Med. 389, 1935–1948 (2023).
Offin, M. et al. Tumor mutation burden and efficacy of EGFR-tyrosine kinase inhibitors in patients with EGFR-mutant lung cancers. Clin. Cancer Res. 25, 1063–1069 (2019).
Borghaei, H. et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N. Engl. J. Med. 373, 1627–1639 (2015).
Nasu, Y. et al. Suicide gene therapy with adenoviral delivery of HSV-tK gene for patients with local recurrence of prostate cancer after hormonal therapy. Mol. Ther. J. Am. Soc. Gene Ther. 15, 834–840 (2007).
Topf, N., Worgall, S., Hackett, N. R. & Crystal, R. G. Regional ‘pro-drug’ gene therapy: intravenous administration of an adenoviral vector expressing the E. coli cytosine deaminase gene and systemic administration of 5-fluorocytosine suppresses growth of hepatic metastasis of colon carcinoma. Gene Ther. 5, 507–513 (1998).
Patel, P. A phase I/II clinical trial in localized prostate cancer of an adenovirus expressing nitroreductase with CB1954 [correction of CB1984]. Mol. Ther. J. Am. Soc. Gene Ther. 17, 1292–1299 (2009).
Rainov, N. G. A phase III clinical evaluation of herpes simplex virus type 1 thymidine kinase and ganciclovir gene therapy as an adjuvant to surgical resection and radiation in adults with previously untreated glioblastoma multiforme. Hum. Gene Ther. 11, 2389–2401 (2000).
Sangro, B. et al. A phase I clinical trial of thymidine kinase-based gene therapy in advanced hepatocellular carcinoma. Cancer Gene Ther. 17, 837–843 (2010).
Sagara, T. et al. Successful gene therapy requires targeting the vast majority of cancer cells. Cancer Biol. Ther. 21, 946–953 (2020).
Windbichler, N. et al. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature 473, 212–215 (2011).
Allen, G. M. & Lim, W. A. Rethinking cancer targeting strategies in the era of smart cell therapeutics. Nat. Rev. Cancer 22, 693–702 (2022).
Zhang, J., Kale, V. & Chen, M. Gene-directed enzyme prodrug therapy. AAPS J. 17, 102–110 (2014).
ElOjeimy, S. et al. FasL gene therapy: a new therapeutic modality for head and neck cancer. Cancer Gene Ther. 13, 739–745 (2006).
Touraine, R. L., Ishii-Morita, H., Ramsey, W. J. & Blaese, R. M. The bystander effect in the HSVtk/ganciclovir system and its relationship to gap junctional communication. Gene Ther. 5, 1705–1711 (1998).
Rollins, C. T. et al. A ligand-reversible dimerization system for controlling protein–protein interactions. Proc. Natl Acad. Sci. USA 97, 7096–7101 (2000).
Di Stasi, A. et al. Inducible apoptosis as a safety switch for adoptive cell therapy. N. Engl. J. Med. 365, 1673–1683 (2011).
Iuliucci, J. D. et al. Intravenous safety and pharmacokinetics of a novel dimerizer drug, AP1903, in healthy volunteers. J. Clin. Pharmacol. 41, 870–879 (2001).
Jura, N. et al. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell 137, 1293–1307 (2009).
Lin, J. H. Pharmacokinetic and pharmacodynamic variability: a daunting challenge in drug therapy. Curr. Drug Metab. 8, 109–136 (2007).
Subbiah, V. et al. Pan-cancer efficacy of pralsetinib in patients with RET fusion-positive solid tumors from the phase 1/2 ARROW trial. Nat. Med. 28, 1640–1645 (2022).
Subbiah, V. et al. Structural basis of acquired resistance to selpercatinib and pralsetinib mediated by non-gatekeeper RET mutations. Ann. Oncol. 32, 261–268 (2021).
Cloughesy, T. F. et al. Durable complete responses in some recurrent high-grade glioma patients treated with Toca 511 + Toca FC. Neuro Oncol. 20, 1383–1392 (2018).
Pandha, H. S. et al. Genetic prodrug activation therapy for breast cancer: a phase I clinical trial of erbB-2-directed suicide gene expression. J. Clin. Oncol. 17, 2180–2189 (1999).
Vermes, A., Guchelaar, H.-J. & Dankert, J. Flucytosine: a review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions. J. Antimicrob. Chemother. 46, 171–179 (2000).
Longley, D. B., Harkin, D. P. & Johnston, P. G. 5-Fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer 3, 330–338 (2003).
Fuchita, M. et al. Bacterial cytosine deaminase mutants created by molecular engineering show improved 5-fluorocytosine-mediated cell killing in vitro and in vivo. Cancer Res. 69, 4791–4799 (2009).
Pardini, B. et al. 5-Fluorouracil-based chemotherapy for colorectal cancer and MTHFR/MTRR genotypes. Br. J. Clin. Pharmacol. 72, 162–163 (2011).
Gilad, Y., Gellerman, G., Lonard, D. M. & O’Malley, B. W. Drug combination in cancer treatment—from cocktails to conjugated combinations. Cancers 13, 669 (2021).
Freytag, S. O. et al. Phase I study of replication-competent adenovirus-mediated double-suicide gene therapy in combination with conventional-dose three-dimensional conformal radiation therapy for the treatment of newly diagnosed, intermediate- to high-risk prostate cancer. Cancer Res. 63, 7497–7506 (2003).
Upadhyay, R. et al. A critical role for fas-mediated off-target tumor killing in T cell immunotherapy. Cancer Discov. 11, 599–613 (2021).
Janopaul-Naylor, J. R., Shen, Y., Qian, D. C. & Buchwald, Z. S. The abscopal effect: a review of pre-clinical and clinical advances. Int. J. Mol. Sci. 22, 11061 (2021).
Roth, J. A. et al. Retrovirus-mediated wild-type P53 gene transfer to tumors of patients with lung cancer. Nat. Med. 2, 985–991 (1996).
Soria, J.-C. et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N. Engl. J. Med. 378, 113–125 (2018).
Kamiyama, D. et al. Versatile protein tagging in cells with split fluorescent protein. Nat. Commun. 7, 11046 (2016).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Dempster, J. M. et al. Extracting biological insights from the Project Achilles genome-scale CRISPR screens in cancer cell lines. Preprint at bioRxiv https://doi.org/10.1101/720243 (2019).
Zeng, H. et al. Genome-wide CRISPR screening reveals genetic modifiers of mutant EGFR dependence in human NSCLC. eLife 8, e50223 (2019).
Maley, C. C., Reid, B. J. & Forrest, S. Cancer prevention strategies that address the evolutionary dynamics of neoplastic cells: simulating benign cell boosters and selection for chemosensitivity. Cancer Epidemiol. Biomark. Prev. 13, 1375–1384 (2004).
Freeman, S. M. et al. The ‘bystander effect’: tumor regression when a fraction of the tumor mass is genetically modified. Cancer Res. 53, 5274–5283 (1993).
Gatenby, R. A., Silva, A. S., Gillies, R. J. & Frieden, B. R. Adaptive therapy. Cancer Res. 69, 4894–4903 (2009).
Zhang, J., Cunningham, J. J., Brown, J. S. & Gatenby, R. A. Integrating evolutionary dynamics into treatment of metastatic castrate-resistant prostate cancer. Nat. Commun. 8, 1816 (2017).
Lin, K. H. et al. Using antagonistic pleiotropy to design a chemotherapy-induced evolutionary trap to target drug resistance in cancer. Nat. Genet. 52, 408–417 (2020).
Zhao, B. et al. Exploiting temporal collateral sensitivity in tumor clonal evolution. Cell 165, 234–246 (2016).
Chen, G. et al. Targeting the adaptability of heterogeneous aneuploids. Cell 160, 771–784 (2015).
Dalin, S., Grauman-Boss, B., Lauffenburger, D. A. & Hemann, M. T. Collateral responses to classical cytotoxic chemotherapies are heterogeneous and sensitivities are sparse. Sci. Rep. 12, 5453 (2022).
Körbelin, J. et al. Pulmonary targeting of adeno-associated viral vectors by next-generation sequencing-guided screening of random capsid displayed peptide libraries. Mol. Ther. 24, 1050–1061 (2016).
Goertsen, D., Goeden, N., Flytzanis, N. C. & Gradinaru, V. Targeting the lung epithelium after intravenous delivery by directed evolution of underexplored sites on the AAV capsid. Mol. Ther. Methods Clin. Dev. 26, 331–342 (2022).
Cheng, Q. et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 15, 313–320 (2020).
Nemunaitis, J. et al. A phase I study of telomerase-specific replication competent oncolytic adenovirus (telomelysin) for various solid tumors. Mol. Ther. J. Am. Soc. Gene Ther. 18, 429–434 (2010).
Liu, X., Ding, J. & Meng, L. Oncogene-induced senescence: a double edged sword in cancer. Acta Pharmacol. Sin. 39, 1553–1558 (2018).
Kim, M.-Y. et al. Tumor self-seeding by circulating cancer cells. Cell 139, 1315–1326 (2009).
Zhu, H. et al. Oncogene-induced senescence: from biology to therapy. Mech. Ageing Dev. 187, 111229 (2020).
Mack, E. T., Perez-Castillejos, R., Suo, Z. & Whitesides, G. M. Exact analysis of ligand-induced dimerization of monomeric receptors. Anal. Chem. 80, 5550–5555 (2008).
Wang, B. et al. Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat. Protoc. 14, 756–780 (2019).
Therneau, T. M. & Grambsch, P. M. Modeling Survival Data: Extending the Cox Model (Springer, 2000).
pritchardlabatpsu/SelectionGeneDrives: Selection Gene Drive Code Release v1.0. GitHub https://doi.org/10.5281/zenodo.10840332 (2024).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR–Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Rodriguez de la Fuente, L., Law, A. M. K., Gallego-Ortega, D. & Valdes-Mora, F. Tumor dissociation of highly viable cell suspensions for single-cell omic analyses in mouse models of breast cancer. STAR Protoc. 2, 100841 (2021).
Leighow, S. M. et al. Programming tumor evolution with selection gene drives to prevent the emergence of drug resistance. Datasets. NCBI Bioproject. NCBI https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1081395 (2024).
Acknowledgements
We acknowledge L. Randolph, V. Rivera, M. Hemann and P. Bruno for their helpful comments on previous versions of the manuscript. We also thank the members of the U01 Synthetic Biology in Cancer consortium for valuable comments during the preparation of the manuscript. We also acknowledge the Huck Flow Cytometry Facility and its members, including D. R. Abrams, M. Koptchak and R. Mani. J.R.P. is supported by U01CA265709, R21EB026617, NSF RECODE CBET 2033673 and NSF Modulus MCB 2141650. This project was supported by Huck Institutes of the Life Sciences at Penn State University through the Huck Innovative and Transformational Seed Grant (HITS). Content is the responsibility of the authors and does not represent the views of the Huck Institutes.
Author information
Authors and Affiliations
Contributions
S.M.L. and J.R.P. conceptualized this work. S.M.L., D.W. and J.R.P. developed the theoretical models. S.M.L., J.A.R., I.S., Z.Y. and H.I. conducted the experiments. S.Y. and M.A. provided support for in vivo studies. S.M.L. wrote the initial draft, and J.R.P. edited the manuscript. J.R.P. acquired the funds to support the project.
Corresponding author
Ethics declarations
Competing interests
J.R.P. is a co-founder of Theseus Pharmaceuticals and holds equity in Theseus Pharmaceuticals. J.R.P. consults for and holds equity in MOMA Therapeutics. J.R.P. and S.M.L. are co-founders of Red Ace Bio. J.R.P. and S.M.L. have filed for patent protection of the work described in the manuscript.
Peer review
Peer review information
Nature Biotechnology thanks Andriy Marusyk and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–7 and Appendices 1 and 2.
Supplementary Video 1
Example spatial agent-based model simulation for small kill radius (ρ = 1) and low dispersion (high initial compactness, θ = 1).
Supplementary Video 2
Example spatial agent-based model simulation for small kill radius (ρ = 1) and high dispersion (low initial compactness, θ = 0).
Supplementary Video 3
Example spatial agent-based model simulation for large kill radius (ρ = 5) and low dispersion (high initial compactness, θ = 1).
Supplementary Video 4
Example spatial agent-based model simulation for large kill radius (ρ = 5) and high dispersion (low initial compactness, θ = 0).
Source data
Source Data Fig. 1
Full scan for blot in Fig. 2e (low exposure time).
Source Data Fig. 2
Full scan for blot in Fig. 2e (high exposure time). Membrane slice containing GAPDH control was not included to avoid overexposure.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Leighow, S.M., Reynolds, J.A., Sokirniy, I. et al. Programming tumor evolution with selection gene drives to proactively combat drug resistance. Nat Biotechnol 43, 737–751 (2025). https://doi.org/10.1038/s41587-024-02271-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-024-02271-7
This article is cited by
-
Erlotinib-Conjugated O-Phenylenediamine Derived Fluorescent Carbon Nanodots (OPD@CD) Platform for Targeting Breast Cancer (MCF-7) Cells
Journal of Fluorescence (2025)
-
Pre-empting drug resistance
Nature Reviews Cancer (2024)